4. Fed Batch Bioreactor
This model represents the fed batch bioreactor in section 2.4.9 of Seborg et al.
Nr |
Symbol |
Name |
Units |
|---|---|---|---|
1 |
\(\mu_\text{max}\) |
Maximum specific growth rate |
1/hr |
2 |
\(K_s\) |
Monod constant |
g/L |
3 |
\(Y_{X/S}\) |
Cell yield coefficient |
1 |
4 |
\(Y_{P/S}\) |
Product yield coefficient |
1 |
5 |
\(X\) |
Cell mass concentration |
g/L |
6 |
\(S\) |
Substrate mass concentration |
g/L |
7 |
\(V\) |
Volume |
L |
8 |
\(P\) |
Substrate mass concentration |
g/L |
9 |
\(F\) |
Feed flow rate |
L/hr |
10 |
\(S_f\) |
Substrate mass concentration |
g/L |
11 |
\(\mu\) |
Specific growth rate |
1/hr |
12 |
\(r_g\) |
Cell growth rate |
g /( L hr) |
13 |
\(r_p\) |
Product growth rate |
g /( L hr) |
Model equations:
Nr |
Equation |
Inputs |
Outputs |
Parameters |
|---|---|---|---|---|
1 |
\[r_g = \mu X\]
|
\(r\), \(\mu\), \(X\) |
||
2 |
\[\mu = \mu_{max} \frac{S}{K_S + S}\]
|
\(S\) |
\(\mu_\text{max}\), \(K_s\) |
|
3 |
\[r_p = Y_{P/X}r_G\]
|
\(r_p\) |
\(Y_{P/X}\) |
|
4 |
\[\frac{d(XV)}{dt} = Vr_g\]
|
\(V\) |
||
5 |
\[\frac{d(PV)}{dt} = Vr_p\]
|
\(P\) |
||
6 |
\[\frac{d(SV)}{dt} = FS_f - Vr\]
|
\(F\), \(S_f\) |
\(Y_{X/S}\) |
|
7 |
\[\frac{dV}{dt} = F\]
|
|||
Number |
7 |
2 |
7 |
4 |
Parameters
[1]:
mu_max = 0.2 # Maximum growth rate
K_s = 1.0 # Monod constant
Y_xs = 0.5 # Cell yield coefficient
Y_px = 0.2 # Product yield coefficient
States
[2]:
X = 0.05 # Concentration of the cells
S = 10 # Concentration of the substrate
P = 0 # Concentration of the product
V = 1 # Reactor volume
x0 = [X, S, P, V] # State vector
Inputs
[3]:
F = 0.05 # Feed rate
S_f = 10 # Concentration of substrate in feed
[4]:
def dxdt(t, x):
[X, S, P, V] = x
mu = mu_max * S / (K_s + S)
rg = mu * X
rp = Y_px * rg
dVdt = F
dXdt = 1/V*(V * rg - dVdt*X)
dPdt = 1/V*(V * rp - dVdt*P)
dSdt = 1/V*(F * S_f - 1 / Y_xs * V * rg - dVdt*S)
return [dXdt, dSdt, dPdt, dVdt]
[5]:
import scipy.integrate
import numpy
[6]:
import matplotlib.pyplot as plt
%matplotlib inline
[7]:
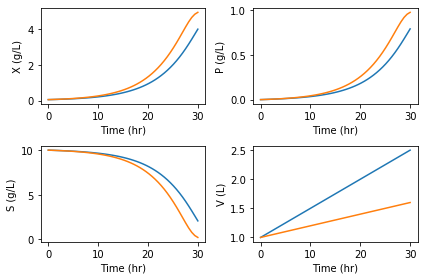
tspan = [0, 30]
[8]:
tsmooth = numpy.linspace(0, 30)
[9]:
Fs = [0.05, 0.02]
[10]:
results = []
[11]:
for F in Fs:
out = scipy.integrate.solve_ivp(dxdt, tspan, x0, t_eval=tsmooth)
results.append(out)
[12]:
names = ['X', 'S', 'P', 'V']
units = {'X': 'g/L', 'S': 'g/L', 'P': 'g/L', 'V': 'L'}
[13]:
ax = {}
fig, [[ax['X'], ax['P']], [ax['S'], ax['V']]] = plt.subplots(2, 2)
for F, out in zip(Fs, results):
var = {name: y for name, y in zip(names, out.y)}
for name in names:
ax[name].plot(out.t, var[name])
ax[name].set_ylabel(f'{name} ({units[name]})')
ax[name].set_xlabel('Time (hr)')
plt.tight_layout()
[ ]: