This notebook times a couple of ways of integrating a number of tanks in series
[1]:
import matplotlib.pyplot as plt
%matplotlib inline
[2]:
Ntimes = 1000
Nstates = 20
t_end = 100
D = 2
[3]:
import numpy
1. No delays
1.1. Normal way
Use a growing list for the history and an array for states
[4]:
ts = numpy.linspace(0, t_end, Ntimes)
dt = ts[1]
[5]:
Fin = 2
k = 1
A = 1
[6]:
%%time
statehistory = []
states = numpy.ones(Nstates)
for i, t in enumerate(ts):
Fout = states[0]*k
dsdt = [1/A*(Fin - Fout)]
for j in range(1, Nstates):
Fin_j = k*states[j - 1]
Fout_j = k*states[j]
dsdt.append(1/A*(Fin_j - Fout_j))
states += numpy.array(dsdt)*dt
# we have to copy because the above += is in place.
statehistory.append(states.copy())
Wall time: 32 ms
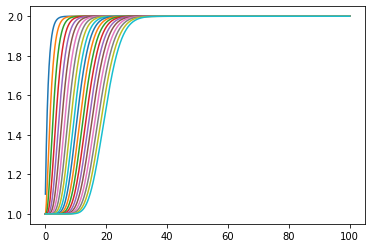
[7]:
plt.plot(ts, statehistory);
1.2. Preallocation
If you are used to Matlab you may imagine pre-allocating statehistory would save lots of time
[8]:
%%time
statehistory = numpy.empty((Ntimes, Nstates))
states = numpy.ones(Nstates)
for i, t in enumerate(ts):
Fout = states[0]*k
dsdt = [1/A*(Fin - Fout)]
for j in range(1, Nstates):
Fin_j = k*states[j - 1]
Fout_j = k*states[j]
dsdt.append(1/A*(Fin_j - Fout_j))
states += numpy.array(dsdt)*dt
statehistory[i, :] = states
Wall time: 32 ms
[9]:
plt.plot(ts, statehistory);
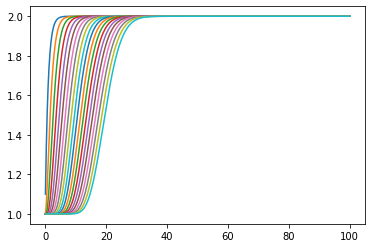
Same result, much the same amount of time.
2. Dead time
Now, let’s introduce a delay between each tank.
2.1. Lists and interp
[10]:
%%time
statehistory = [[] for _ in states]
states = numpy.ones(Nstates)
for i, t in enumerate(ts):
Fout = states[0]*k
dsdt = [1/A*(Fin - Fout)]
for j in range(1, Nstates):
delayed_Fin_j = k*(numpy.interp(t - D, ts[:i], statehistory[j-1]) if t > 0 else states[j-1])
Fout_j = k*states[j]
dsdt.append(1/A*(delayed_Fin_j - Fout_j))
states += numpy.array(dsdt)*dt
for j, s in enumerate(states):
statehistory[j].append(s)
Wall time: 639 ms
[11]:
plt.plot(ts, numpy.array(statehistory).T);
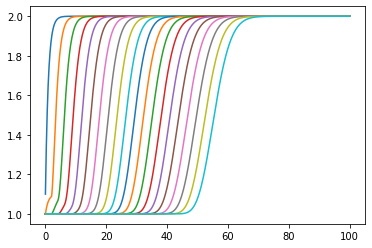
OK, that took a lot longer.
2.2. Approximate indexing
What if we just use indexing instead of interpolation?
[12]:
%%time
statehistory = [[] for _ in states]
states = numpy.ones(Nstates)
for i, t in enumerate(ts):
Fout = states[0]*k
dsdt = [1/A*(Fin - Fout)]
for j in range(1, Nstates):
delayed_Fin_j = k*(statehistory[j-1][i - int(D/dt)] if t > D else states[j-1])
Fout_j = k*states[j]
dsdt.append(1/A*(delayed_Fin_j - Fout_j))
states += numpy.array(dsdt)*dt
for j, s in enumerate(states):
statehistory[j].append(s)
Wall time: 49 ms
OK, we’re back to almost the same time as before, but do we get the same result?
[13]:
plt.plot(ts, numpy.array(statehistory).T);
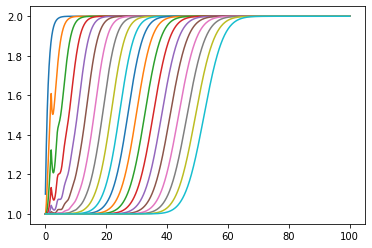
No. the rounding errors build up. If we use this strategy we had better choose a step size which divides cleanly into the dead time.